
Figure legend: Role of the GVCC and synergy with EpiMVP projects and the HETC
Milestone 1: Selection of genes and variants for study in EpiMVP projects 1-3.
- Gene selection: Although there are hundreds of genes implicated in epilepsy, the GVCC will select genes for study by EpiMVP based on a number of criteria.
- Variant selection: Both pathogenic missense variants (positive controls) and VUS will be collated from patients (with industry partners) and benign variants from population genetic sequencing datasets.

Fig 2. Distributions of pathogenic variants in known epilepsy genes The top 20 genes implicated in epilepsy in 18,705 patients. Non-ion-channel, non-ligand gated channel genes that are candidates for study in EpiMVP are shown in purple.
Milestone 2: Develop EpiPred, a novel epilepsy-specific computational predictive model, using EpiMVP Project-generated and existing functional data to accurately predict the likelihood of a variant being associated with epilepsy.
We’ll use a variety of resources to characterize variants, including:
- Existing variant interpretation annotation tools
- Proteomics and structural modeling
- EpiMVP functional data
EpiPred for variant classification and prioritization will be devised following an iterative machine learning model. Annotated variant data will be used as input to EpiPred for prioritization of missense variants predicted to be pathogenic, VUS or benign. This will aid functional characterization in projects 1-3, and functional data will be used as input in the model. In this iterative process, increasing levels of complex functional data will be used to hone the accuracy of EpiPred to classify variants.
Milestone 3: To support data management and web-based resources needed for seamless data sharing and implementation of EpiPred in the epilepsy community.
- The GVCC will co-ordinate data from all participating insititutaions.
- EpiPred will eventually have a web-facing version that will allow users (clinical care providers and patients/families) to generate prediction on their own variants.
- We will also work with ClinGen, ClinVar, the ACMG and industry partners to integrate the prediction score into standard genetic testing results.
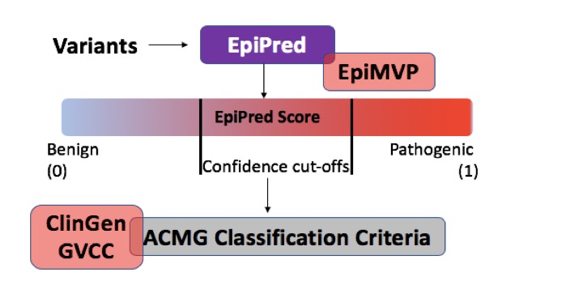
EpiPred classification and integration
